From 2011 through 2021, we served as the coordinating center (CC) for NCI's Barrett’s Esophagus Translational Research Network (BETRNet), a consortium of research centers working in close scientific partnership with one another and NCI program staff to energize and synergize investigation into the underlying biology of esophageal adenocarcinoma (EAC), a malignancy whose incidence in the United States has risen sharply in recent decades. In particular, BETRNet's participants were motivated to apply innovative multi-institutional and transdisciplinary research approaches to uncover the biology of EAC and Barrett’s esophagus (BE), the only known EAC precursor lesion, with the end goal of validated strategies for patient management, resulting in improved outcomes.
As BETRNetCC, we implemented a clear administrative and scientific vision for coordinating center operations in support of BETRNet goals, including:
- coordination and facilitation
- organization and support for the pilot project program, including data analysis and results interpretation
- scientific collaboration and contribution to novel and innovative methodologies
- development, maintenance, and administration of the BETRNet patient registry with virtual biorepository (PR-VB), built with novel informatics technologies and machine learning–based search tools
The result was ten years of efficient, effective coordination and facilitation of BETRNet meetings, teleconferences, and communications.
Coordination and facilitation
Annual face-to-face meetings
Throughout the BETRNet initiative, the most intensive promotion of collaboration, cooperation, and building synergies took place during the annual investigator meetings, organized and coordinated by BETRNetCC in partnership with NCI staff. The logistical demands of planning and executing each meeting included the following:
- polling steering committee members to identify mutually available dates for the two-day meeting, with direction from NCI staff
- coordinating the reservation of meeting spaces and hotel rooms, in collaboration with local planners. (The in-person meetings were held at non-VUMC sites.)
- collecting attendee information in advance to facilitate secure check-in, taxi and other transportation arrangements, and audiovisual configurations, and address other needs
- collecting and organizing presentation slides
- preparing the meeting agenda, roster, and name tags, in coordination and collaboration with NCI program staff and research center PIs
- coordinating travel and reimbursements for invited guest speakers
- facilitating the proceedings as they were conducted, including set-up and support of remote access for participants unable to attend in person (and, in 2020 and 2021, remote access for all participants because of COVID-19 precautions)
- taking meeting minutes and keeping time
- organizing an on-site dinner to encourage more casual social interaction among BETRNet participants
- distributing minutes and following up on other action items after the meeting
Each meeting consisted of one day for scientific presentations, including presentation of the accomplishments of each of the research centers to date, as well as talks by invited guest speakers on topics of special interest in BE/EAC; the second day of each meeting is a half-day business meeting. (Note: The Y1 meeting was an exception)
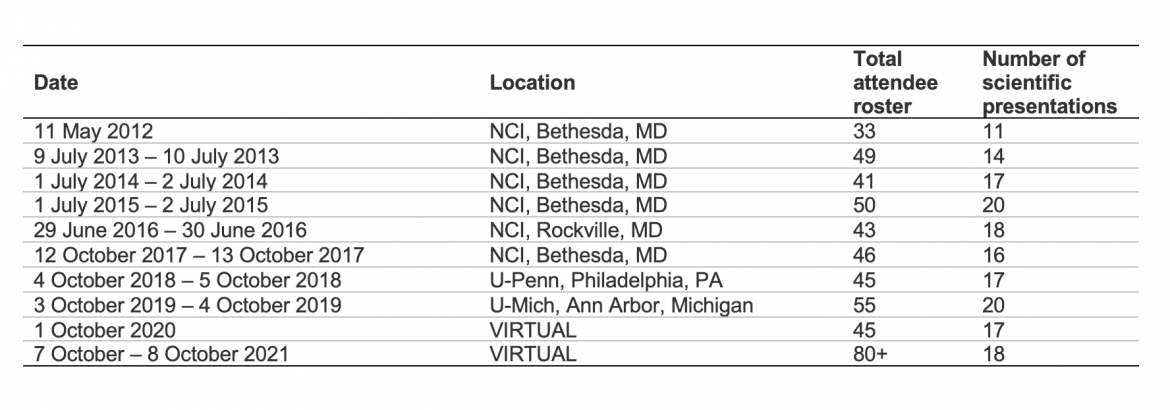
This table provides a summary of the annual meetings.
Periodic business meetings and teleconferences
In addition to organizing and coordinating the annual meetings, BETRNetCC, in collaboration with NCI program staff and Steering Committee co-chairs, organized and hosted periodic business meetings to facilitate routine and ongoing communication, as well as to continue promoting collaborative efforts among all BETRNet members.
BETRNetCC activities related to these teleconferences included:
• identifying teleconference dates with maximum availability of meeting participants
• preparing and distributing meeting agendas and any auxiliary meeting materials
• providing meeting reminders
• setting up the technology needed for these meetings, such as teleconference lines or web conferencing applications
• facilitating and leading the teleconference
• preparing and distributing meeting minutes, and following up on action items
The meetings covered topics such as:
• cross-BETRNet pilot project initiation, progress, and final paper submission
• other group paper plans and progress
• visibility of BETRNet research at scientific meetings
• progress of the BETRNet PR-VB (see below for details)
• other administrative and business items
Scientific seminars
In year 3 of BETRNet, BETRNetCC assumed responsibility for organizing and coordinating scientific web seminars for program members. These conferences—a total of 18 distinct talks—offered opportunities for individual BETRNet investigators, as well as BE/EAC researchers outside the network, to provide in-depth presentations of their research, followed by discussion among conference participants. The speakers and presentations are listed on our BETRNet seminars page.
BETRNetCC activities related to these webinars included:
• setting the schedule
• preparing and distributing topic introductions for the speaker/talk ahead of each seminar
• providing seminar reminders
• setting up web conferencing, which included meeting with speakers ahead of time to ensure their facility with the web conferencing software features
• gathering speaker slides and other materials to be presented in the seminar
• moderating the seminar, including keeping time and managing the question-and-answer portion
Pilot project program
As a mechanism to make the most of opportunities for collaboration, integrating and coordinating research activities across all BETRNet research centers, a cross-BETRNet pilot project was featured during each grant cycle. The first cross-BETRNet pilot project focused on the discovery and validation of biomarkers in relation to high and low risk of progression on the BE spectrum; during the second cycle, the focus was on genomic and epigenomic drivers of persistent and recurring BE.
For the pilot project program, BETRNetCC provided:
- administrative support for the proposal process. In consultation with NCI staff, BETRNetCC developed and distributed timelines and requirements for the process, and assisted with the collection and administrative review of proposal materials.
- organization, hosting, and facilitation of cross-BETRNet conferences at each stage of the project.
- In an initial conference during each grant cycle, all the research center investigators discussed opportunities for a meaningful pilot project, with the goal of leveraging their combined resources to accomplish specific aims that would not be possible within the resource constraints of a single institution.
- With an opportunity identified and agreed on, a work group was tasked with translating it into a detailed research proposal to be submitted to NCI. BETRNetCC coordinated the teleconferences for this group.
- Upon NCI approval, BETRNetCC convened conferences for the pilot project site PIs designated by each research center, during which timelines were established, progress was reviewed, challenges underwent troubleshooting, and other tasks performed to continue moving the project forward
- Finally, BETRNetCC organized teleconferences for discussion and review of the final paper to be published with project findings.
- data management. To collect the specific demographic, exposure, and clinical covariates of interest for each study, BETRNetCC worked with investigators to determine the relevant covariates and develop a set of data fields harmonized across the research centers. For pilot project 2, BETRNetCC further constructed a project-specific REDCap database to facilitate submission of the common data elements. The database also made it possible to track tissue specimens to be utilized in the study, from approval and shipment to central pathology, to receipt and DNA isolation by central path, to distribution of DNA for sequencing and methylation analysis.
- data analysis. For pilot project 1, research centers submitted experimental data to BETRNetCC, and we performed a series of analyses to evaluate and validate the performance of the selected biomarkers, using simple logistic regression as well as five different machine learning methods. For pilot project 2, data analysis is pending.
Patient registry with virtual biorepository
A major function of BETRNetCC was developing and implementing the BETRNet patient registry with virtual biorepository (BETRNet PR-VB), to support data and specimen sharing across the network. This involved the following key activities.
Identification of a minimum dataset (MDS)
In developing the virtual biorepository, a critical first step was to define a BETRNet-specific minimum dataset—that is, key clinic-pathologic features to include in the patient registry that would allow for identification of patient subgroups and biospecimens of interest. This would optimize the use of these resources to answer research questions. To address this need, BETRNetCC convened the BETRNet Minimum Dataset Working Group (MDWG), with its members bringing clinical and informatic expertise from across the BETRNet.
At the time, a number of long-standing BE/EAC registries with extensive data existed across the network, but each local database varied somewhat from the others in terms of the structure and content of data elements collected. Therefore, the MDWG was further charged with establishing common data elements (CDEs) for the MDS, such that local data elements would be mapped to variables with common definitions and understanding of terms across the network.
Launch of the PR-VB
After specification of the MDS, BETRNetCC proceeded with development of the PR-VB; the system was designed and launched within three months of the MDS's finalization.
Convening of the virtual biorepository working group (VBWG)
With the MDS finalized and the PR-VB launched, the MDWG was dissolved. BETRNetCC then convened the VBWG to guide and support efforts related to data and biospecimen collection, submission, and tracking in the PR-VB. BETRNetCC coordinated and hosted multiple teleconferences for this group to address and resolve technical implementation issues related to data submission, as well as to develop solutions for challenges related to informed consent (in conjunction with how the data and specimens originally had been collected), along with regulatory requirements.
Support for data submissions
With the PR-VB launched into production, BETRNetCC turned its attention to facilitating data submissions. This included troubleshooting technical problems and rapidly implementing modifications as needed to remove system-side obstacles, such as:
- modifications to both allowable values for categorical variables and data validations to account for edge cases encountered during data submissions
- modifications to the data validation error report in response to user input on how to improve the report to better support user ability to understand and correct errors as flagged
- modifications to the BETRNet patient identifier assignment algorithm to address edge cases associated with incremental submission of data associated with the same subject
Specimen access protocol
In collaboration with NCI program staff, and in accordance with NCI Best Practices for Biospecimen Resources, BETRNetCC drafted a protocol for accessing PR-VB data and requesting specimens; this was reviewed by members of the BETRNet Steering Committee, with subsequent final review by an expert in bioethics. Upon finalization of the protocol, BETRNetCC convened a specimen access review committee, charged with reviewing and scoring access proposals. It consisted of representatives from each BETRNet research center, plus an NCI expert in bioethics.
Specimen request infrastructure
Following development of the data capture and management portion of the BETRNet PR-VB, BETRNetCC software development efforts focused on web-based infrastructure to support the specimen request process. The web portal for this purpose was developed as two additional modules within the PR-VB: (1) the My Projects module, for investigators to build and track request proposals, and (2) the Specimen Distribution module, for biobanks to receive specimen distribution requests and record their fulfillment of these requests. Key features of these modules included automation of all communications throughout the specimen request and fulfillment process, automated tracking of the status of each request, and use of a project file log to store all files passed through the system, including the access proposal, list of requested specimens, list of requested data elements, review committee comments on the proposal, data file, any relevant material transfer agreements (MTAs) between the investigator's institution and the biobank, specimen shipment manifest files, and so on.
Scientific collaboration and contribution to novel and innovative methodologies
Throughout the lifetime of the BETRNet program, BETRNetCC developed and advanced methodologies for analyzing and interpreting the types of complex genomic data generated in various BETRNet research projects. These methodologies have been applied directly to collaborations with BETRNet research centers, leading to collaborative publications with BETRNet investigators (Liu et al., Biomed Res Int 2015; Blum et al. Gastroenterology 2019; Blum et al. Cancer Res 2016), as well as numerous method publications, with Yu Shyr as senior author. Here are some of the methodologies developed and/or applied in BETRNet collaborations:
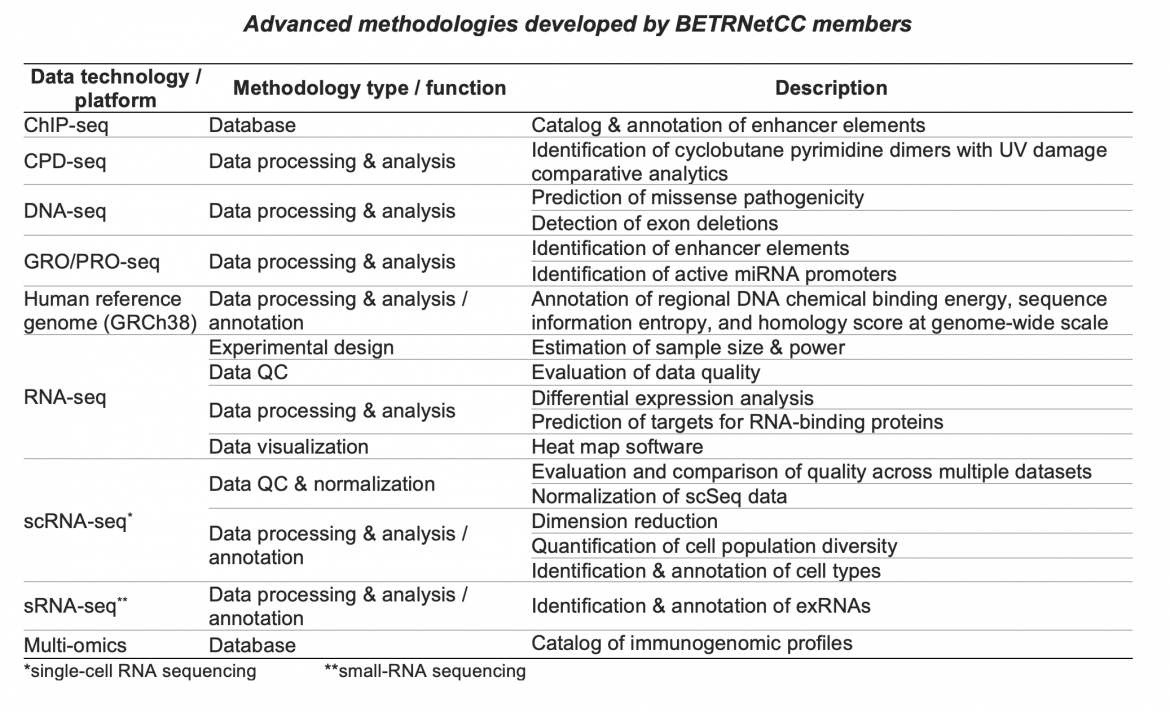
Click the References link below to view publication details. See also the CQS Software Tools page.
-
Collaborative BETRNet publications
Liu Q, Zhong X, Madison BB, Rustgi AK, Shyr Y. Assessing computational steps for CLIP-seq data analysis. Biomed Res Int. 2015;2015:196082.
Blum AE, Venkitachalam S, Ravillah D, et al. Systems biology analyses show hyperactivation of transforming growth factor-β and JNK signaling pathways in esophpgeal cancer. Gastroenterology. 2019;156(6):1761-1774.
Blum AE, Venkitachalam S, Guo Y, et al. RNA sequencing identifies transcriptionally viable gene fusions in esophageal adenocarcinomas. Cancer Res. 2016;76(19):5628-5633.
ChIP-seq. Wang J, Dai X, Berry LD, Cogan JD, Liu Q, Shyr Y. HACER: an atlas of human active enhancers to interpret regulatory variants. Nucleic Acids Res. 2019;47:D106-D112.
CPD-seq. Sheng Q, Yu H, Duan M, et al. A streamlined solution for processing, elucidating and quality control of cyclobutane pyrimidine dimer sequencing data. Nat Protoc. 2021;16(4):2190-2212.
DNA-seq. Chen HC, Wang J, Liu Q, Shyr Y. A domain damage index to prioritizing the pathogenicity of missense variants. Hum Mutat. Published online August 5, 2021. doi:10.1002/humu.24269
Guo Y, Zhao S, Lehmann BD, et al. Detection of internal exon deletion with ExonDel. BMC Bioinformatics. 2014;15(1):332.
GRO/PRO-seq. Wang J, Zhao Y, Zhou X, Hiebert SW, Liu Q, Shyr Y. Nascent RNA sequencing analysis provides insights into enhancer-mediated gene regulation. BMC Genomics. 2018;19:633.
Liu Q, Wang J, Zhao Y, et al. Identification of active miRNA promoters from nuclear run-on RNA sequencing. Nucleic Acids Res. 2017;45:e121.
Human reference genome. Sheng Q, Yu H, Oyebamiji O, et al. AnnoGen: annotating genome-wide pragmatic features. Bioinformatics. 2020;36(9)2899-2901.
RNA-seq. Zhao S, Li CI, Guo Y, Sheng Q, Shyr Y. RnaSeqSampleSize: real data based sample size estimation for RNA sequencing. BMC Bioinformatics. 2018;19(1):191.
Guo Y, Zhao S, Sheng Q, et al. Multi-perspective quality control of Illumina exome sequencing data using QC3. Genomics. 2014;103(5):323-328.
Guo Y, Zhao S, Ye F, Sheng Q, Shyr Y. MultiRankSeq: multiperspective approach for RNAseq differential expression analysis and quality control. Biomed Res Int. 2014;2014:248090.
Yu H, Wang J, Sheng Q, Liu Q, Shyr Y. beRBP: binding estimation for human RNA-binding proteins. Nucleic Acids Res. 2019;47:e26.
Zhao S, Guo Y, Sheng Q, Shyr Y. Advanced heat map and clustering analysis using heatmap3. Biomed Res Int.2014;2014:986048.
scRNA-seq. Liu Q, Sheng Q, Ping J, et al. scRNABatchQC: multi-samples quality control for single cell RNA-seq data. Bioinformatics. 2019;35:5306-5308.
Chen B, Ramirez-Solano MA, Heiser CN, Liu Q, Lau KS. Processing single-cell RNA-seq data for dimension reduction-based analyses using open-source tools. STAR Protoc. 2021;2:100450.
Liu Q, Herring CA, Sheng Q, et al. Quantitative assessment of cell population diversity in single-cell landscapes. PLoS Biol. 2018;16(10):e2006687.
sRNA-seq. Allen RM, Zhao S, Ramirez Solano MA, et al. Bioinformatic analysis of endogenous and exogenous small RNAs on lipoproteins. J Extracell Vesicles. 2018;7(1):1506198.
Multi-omics. Yang J, Zhao S, Wang J, Sheng Q, Liu Q, Shyr Y. Immu-Mela: An open resource for exploring immunotherapy-related multidimensional genomic profiles in melanoma. J Genet Genomics. 2021;48:361-368.